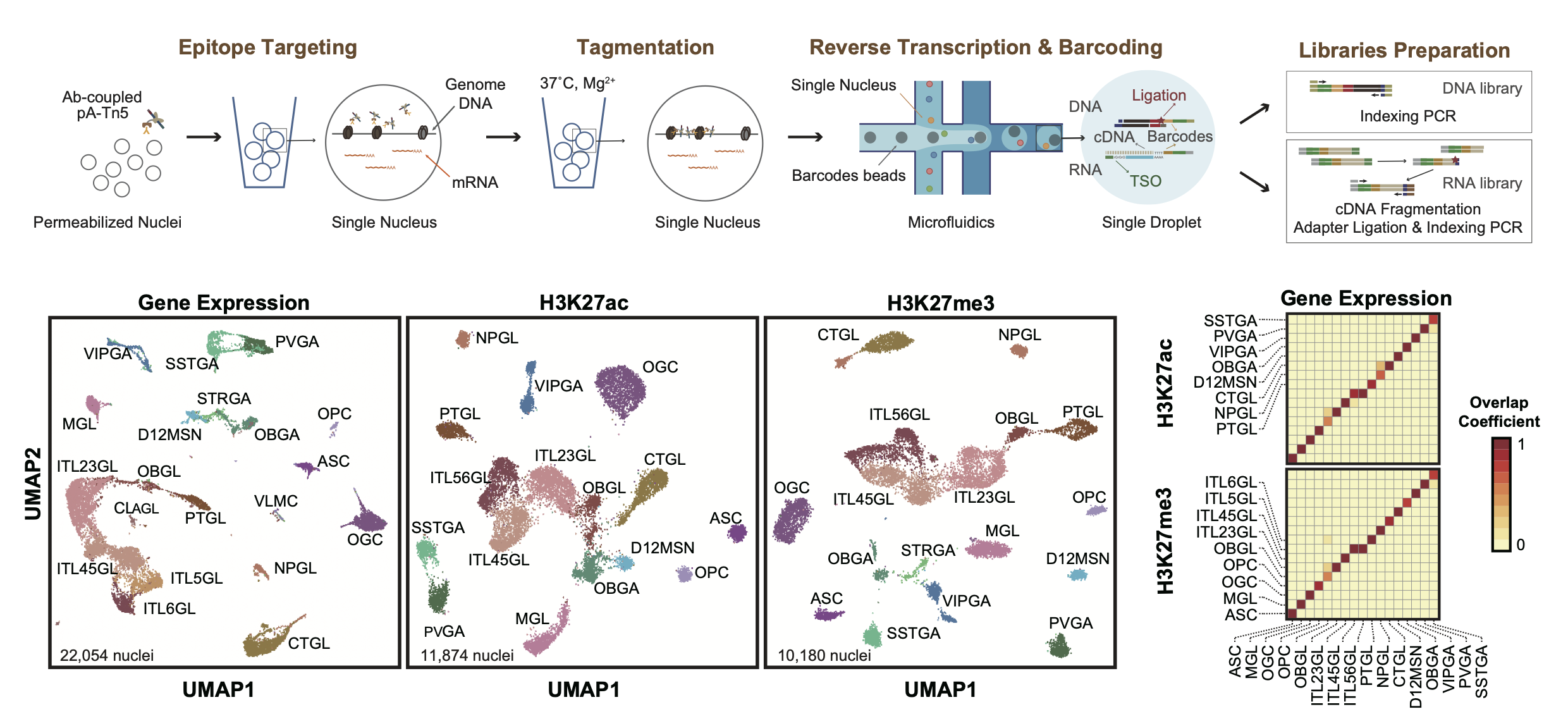
Droplet Paired-Tag
Single-cell method for joint analysis of histone marks and transcriptome
Droplet Paired-Tag is a convenient and robust method for multi-modal profiling of histone modifications and transcriptome in single cell.
Histone modifications are chemical changes that primarily occur on the tails of histones, which are the scaffold proteins around which DNA is wound. These modifications can alter how tightly or loosely the DNA is wrapped around the histones. When DNA is loosely wound, it becomes more exposed and accessible to various protein machinery; conversely, tightly wound DNA is less accessible. This accessiblity can be used to define the activity of DNA sequence.
Understanding the types of modifications present on genes or regulatory elements is crucial for determining the activity of these sequences. When combined with gene expression measurements, we can better distinguish the functional states of regulatory elements. Obtaining this information in individual cells or specific cell types is particularly important, as it can help identify the key cell population in development or diseases, and tell us which genes and their regulatory elements are more likely to determine the fates of these cells.
Compared with previous methods, Droplet Paired-Tag has higher signal specificty and is way easier to do. In our manuscript, we have used this method to resolve the repressive elements in adult mouse brain, which is difficult to do before. Other study has utilized Droplet Paired-Tag to characrize enhancers in human brain, and concluded that epi-conserved regulatory elements across multiple species are more likely to be active enhancers.
For more details, please refer to our manuscript.