MiniAtlas: a multimodal histone modification and transcriptome atlas for adult human brain
Single-Cell Atlas of Transcription and Chromatin States Reveals Regulatory Programs in the Human Brain
The extraordinary diversity of cell types in the human brain is sustained by complex gene regulatory programs that stabilize cellular identity throughout lifespan. While recent single-cell transcriptomic atlases have cataloged this diversity in detail, the epigenetic mechanisms that establish and maintain these programs have remained difficult to access at scale and resolution.
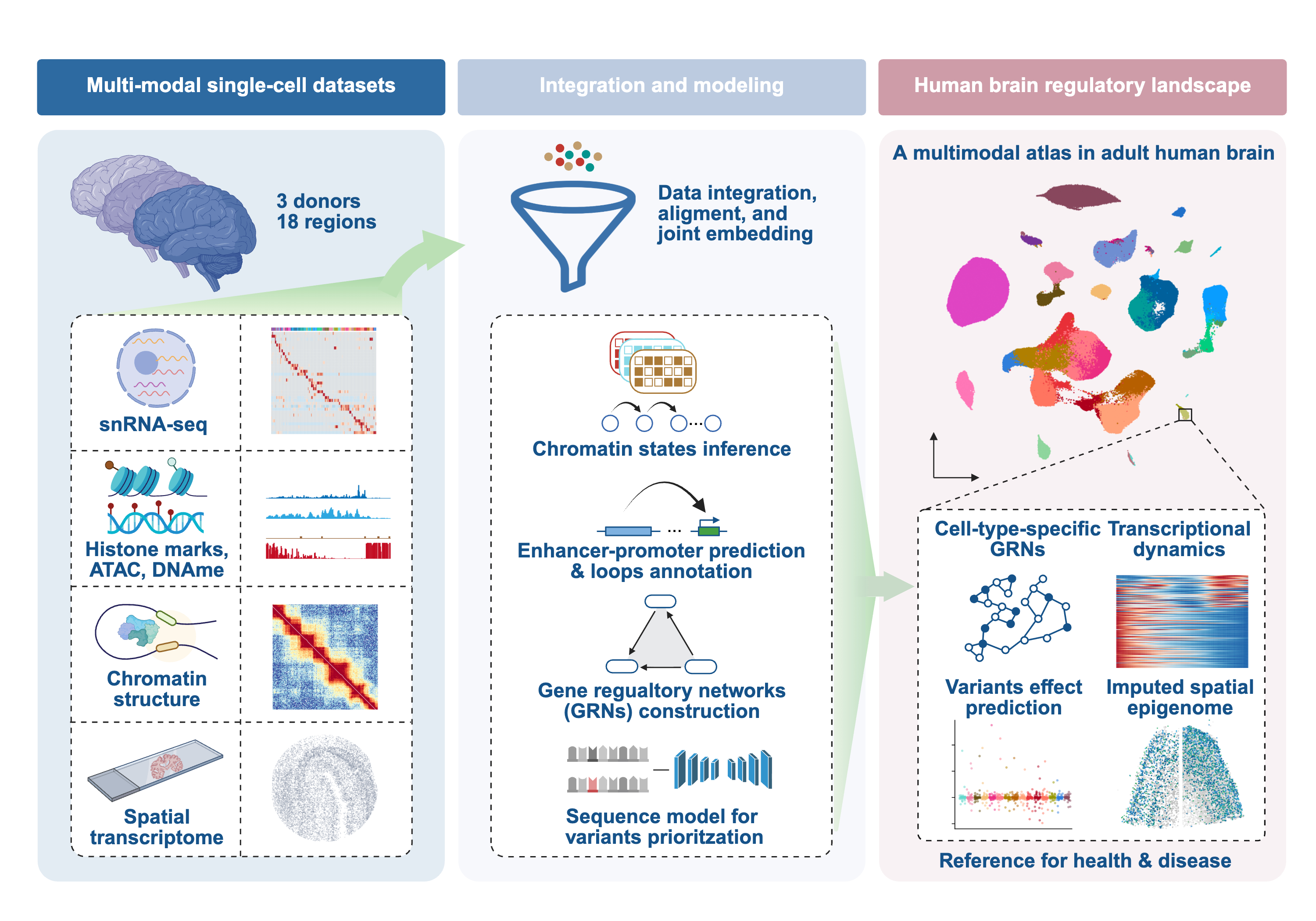
Here, we present a multimodal single-cell atlas jointly profiling gene expression with either active (H3K27ac) or repressive (H3K27me3) histone modifications across more than 750,000 nuclei from 18 adult human brain regions. This resource enables direct interrogation of regulatory activity and organization within a unified cellular framework. Beyond cell type annotation, our study reveals several general principles of neuronal genome regulation:
- Cell-type-resolved chromatin states: We systematically classify the non-coding genome into accessible, active, and repressive states across brain cell types, providing a functional annotation that refines regulatory element catalogs and improves genetic interpretation.
- Prioritization of non-coding disease variants: By resolving cell-type-specific enhancer-promoter links and gene regulatory networks, we connect non-coding neuropsychiatric variants to candidate target genes. By training a sequence-based deep-learning model, we further predict the variant effects on epigenetic signal and gene expression.
- Epigenetic encoding of developmental history: We find that repressive histone modifications preserve signatures of developmental origin and spatial organization. Glial cells retain forebrain-hindbrain repressive signatures, cortical neurons display anterior-posterior gradients in gene activities, and laminar neurons show repressive patterns consistent with their migratory trajectories by our spatial epigenome analysis.
- Neuron-specific long-range repressive interactions: We identify a class of chromatin interactions spanning exceptionally long distance (>5 Mb) that are specific to post-mitotic neurons and mediated by Polycomb-associated repression. These interactions engaged early developmental genes and may contribute to the long-term stabilization of neuronal identity.
- Evolutionary asymmetry in regulatory grammar: Cross-species integration with mouse dataset reveals strong conservation of active transcription factor motif grammar, whereas the repressive grammar is highly divergent, suggesting that evolutionary change in the human brain may preferentially act through silencing mechanisms.
For more details, please refer to our manuscript.