FNIH Heart: dissect regulatory networks underlying heart failures
Single cell multiomics and chromatin architecture reveal human heart failure gene regulatory programs
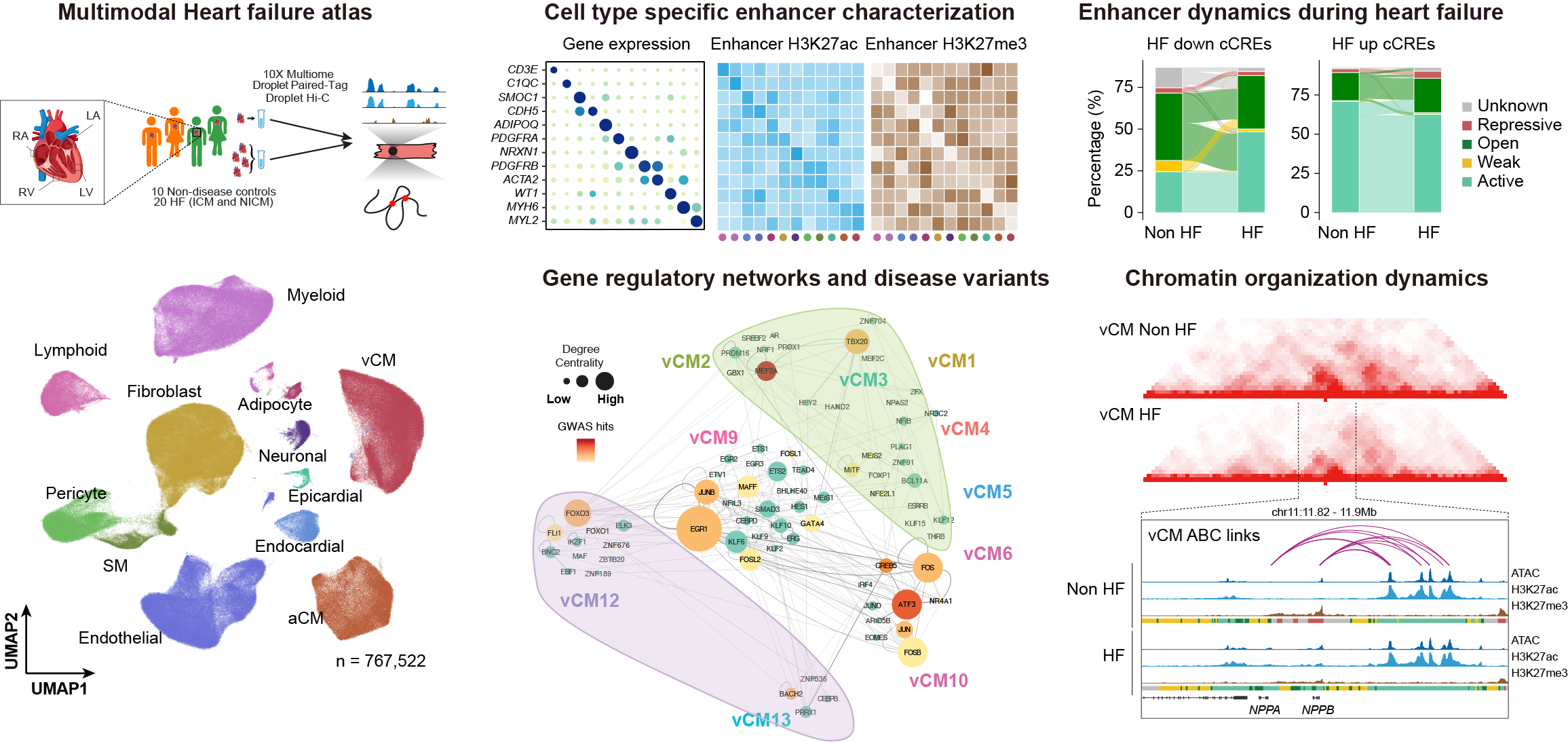
The human heart dynamically adapts to diverse environmental conditions and stresses in health andmdisease; yet how maladaptive gene regulatory responses of specific cardiac cell types contribute to heart failure remains incompletely understood. Here, we present an unprecedented, comprehensive interrogation of cell type-specific gene regulation across 36 human hearts, encompassing non-failing and failing (ischemic and non-ischemic) conditions. Utilizing a novel pooled sample strategy combined with cutting-edge single-cell multimodal assays—including single-cell multiome profiling (10x Chromium single-cell ATAC-seq and RNA-seq), joint single-cell histone modification and gene expression profiling (Droplet Paired-Tag), and single-cell chromatin conformation capture (Droplet Hi-C)—we systematically characterized transcriptional states, chromatin accessibility, histone modifications, and 3D chromatin architecture across all four heart chambers (left and right ventricles and atria).
Our integrative analyses established a detailed, cell-type-resolved atlas of gene regulatory programs and chromatin interactions, dramatically expanding known cardiac cis-regulatory elements by tenfold and revealing novel enhancer-gene interactions specific to distinct cardiac cell types. Comparative analyses between non-failing and failing hearts uncovered significant alterations in cell type composition, gene regulatory landscapes, and global chromatin architecture, with particularly profound disruptions observed in cardiomyocytes and fibroblasts. Strikingly, diseased cardiomyocytes exhibited substantial perturbations in chromatin compartmentalization, providing critical insights into how nuclear remodeling may drive maladaptive gene regulation during heart failure progression.
Our multiomic atlas serves as an invaluable, explorable resource (https://epigenome.wustl.edu/HeartEpigenome/) that systematically links non-coding genetic variants to cell type-specific gene regulatory changes, facilitating the interpretation of variants associated with cardiovascular diseases identified by genome-wide association studies (GWAS) and whole-genome sequencing efforts. Thus, our findings significantly advance the fields of human genetics, chromatin biology, and cardiovascular medicine, providing essential insights and resources that will catalyze the development of precise, targeted therapies for heart failure.
For more details, please refer to our manuscript.